WP 2.1 WEAKLY LABELED LEARNING
With focus on deep learning with large amounts of images that are only weakly labeled (e.g. only overall diagnosis or treatment outcome is available). Techniques were developed that exploit a small set of images with detailed annotations and a large pool of weakly or completely unlabeled data. We exploit shared representations between learning tasks with different localization levels and use active learning where medical experts are asked for feedback on automatically selected cases. We exploit shared representations between learning tasks with different localization levels and use active learning where medical experts are asked for feedback on automatically selected cases.
RESEARCH BY FRISO HESLINGA
We use deep learning techniques for the analysis of ophthalmic images that have been collected by our clinical partners. For example, we work with color fundus photos from Maastricht UMC+ and UMC Utrecht and optical coherence tomography (OCT) scans from Rigshospitalet-Glostrup in Copenhagen. We aim to find biomarkers related to type 2 diabetes in fundus images of the retina and directly predict disease stage. Similarly, we use fundus images of premature infants to predict retinopathy of prematurity. Besides the retina, we also focus on the cornea. OCT images of the anterior segment of the eye are used to find biomarkers related to Descemet's membrane endothelial keratoplasty. Our colleagues at the Medical Image Analysis Group at TU/e have developed deep learning tools for a wide range of applications and medical image types, so there is always someone around with suggestions how to tackle a new challenge. Every now and then we meet with our partners at Philips Research to exchange research ideas and discuss the latest innovations.
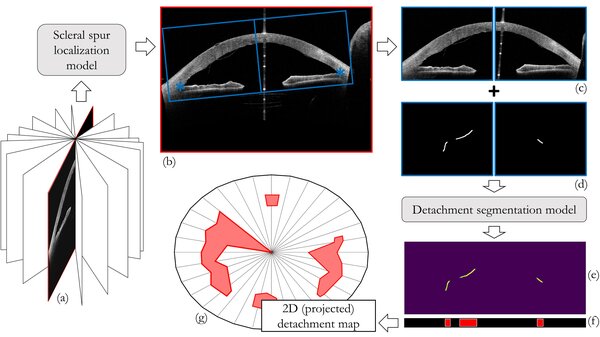
RESEARCH BY SUZANNE WETSTEIN
This project is on the application of deep learning algorithms to histopathological image analysis. We have developed a deep learning system to measure Terminal Duct Lobular Unit (TDLU; structures in the breast that produce milk during lactation) involution. TDLU involution has been related to breast cancer risk in previous studies. In another project we developed a deep learning system to grade ductal carcinoma in situ (DCIS; a pre-malignant breast lesion). Our next project focuses on deep learning algorithms for survival prediction of young breast cancer patients.
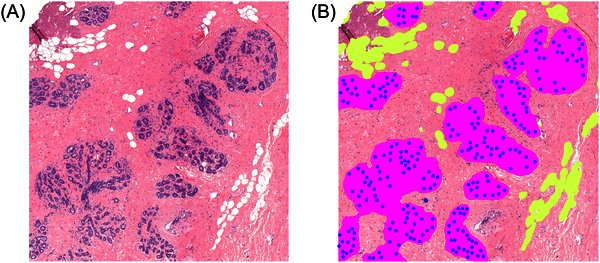
All research projects during my PhD at TU/e were supported by knowledge from Philips. The project in which we developed a deep learning system to measure TDLU involution was a collaboration with the Beth Israel Deaconess Medical Center (BIDMC; Harvard Medical School) in Boston. The system in this project was trained on their data and later also implemented on their servers to be applied to large medical image datasets in future studies. During this project I visited BIDMC for three months and worked closely together with the researchers and pathologists there. The project on the grading of DCIS is in collaboration with the pathology department of the UMC Utrecht and our upcoming project on survival of young breast cancer patients is a collaboration with the NKI and UMC Utrecht. Without the access to these parties within the e/MTIC ecosystem, I would have never been able to perform this research.